Machine learning to segment neutron images
Anders Kaestner, Beamline scientist - Neutron Imaging
Laboratory for Neutron Scattering and Imaging
Paul Scherrer Institut
Lecture outline¶
- Introduction
- Limited data problem
- Unsupervised segmentation
- Supervised segmentation
- Final problem: Segmenting root networks using convolutional NNs
- Future Machine learning challenges in NI
Getting started¶
If you want to run the notebook on your own computer, you'll need to perform the following step:
- You will need to install Anaconda
- Clone the lecture repository (in the location you'd like to have it)
git clone https://github.com/ImagingLectures/MLSegmentation4NI.git
- Enter the folder 'MLSegmentation'
- Create an environment for the notebook
conda env create -f environment. yml -n MLSeg4NI
Enter the environment
conda env activate MLSeg4NI
Start jupyter and open the notebook
lecture/ML4NeutronImageSegmentation.ipynbUse the notebook
Leave the environment
conda env deactivate
Importing needed modules¶
This lecture needs some modules to run. We import all of them here.
import matplotlib.pyplot as plt
import seaborn as sn
import numpy as np
import pandas as pd
import skimage.filters as flt
import skimage.io as io
import matplotlib as mpl
from sklearn.cluster import KMeans
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import confusion_matrix
from sklearn.datasets import make_blobs
from matplotlib.colors import ListedColormap
from matplotlib.patches import Ellipse
from lecturesupport import plotsupport as ps
import scipy.stats as stats
import astropy.io.fits as fits
import keras.metrics as metrics
import keras.losses as loss
import keras.optimizers as opt
from keras.models import Model
from keras.layers import Input, Conv2D, MaxPooling2D, UpSampling2D, concatenate
%matplotlib inline
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('svg', 'png')
#plt.style.use('seaborn')
mpl.rcParams['figure.dpi'] = 300
Using TensorFlow backend.
import importlib
importlib.reload(ps);
Introduction¶
Introduction to neutron imaging
- Some words about the method
- Contrasts
Introduction to segmentation
- What is segmentation
- Noise and SNR
Problematic segmentation tasks
- Intro
- Segmentation problems in neutron imaging
What is an image?¶
A very abstract definition:
- A pairing between spatial information (position)
- and some other kind of information (value).
In most cases this is a two- or three-dimensional position (x,y,z coordinates) and a numeric value (intensity)
Science and Imaging¶
Images are great for qualitative analyses since our brains can quickly interpret them without large programming investements.
Proper processing and quantitative analysis is however much more difficult with images.¶
- If you measure a temperature, quantitative analysis is easy, $T=50K$.
- If you measure an image it is much more difficult and much more prone to mistakes,
- subtle setup variations may break you analysis process,
- and confusing analyses due to unclear problem definition
Furthermore in image processing there is a plethora of tools available¶
- Thousands of algorithms available
- Thousands of tools
- Many images require multi-step processing
- Experimenting is time-consuming
Some word about neutron imaging¶
The transmitted radiation is described by Beer-Lambert's law which in its basic form looks like
$$I=I_0\cdot{}e^{-\int_L \mu{}(x) dx}$$Image types obtained with neutron imaging¶
| Fundamental information | Additional dimensions | Derived information |
|---|---|---|
| 2D Radiography | Time series | q-values |
| 3D Tomography | Spectra | strain |
| Crystal orientation |
Neutron imaging contrast¶
| Transmission through sample | X-ray attenuation | Neutron attenuation |
Measurements are rarely perfect¶
Factors affecting the image quality¶
- Resolution (Imaging system transfer functions)
- Noise
- Contrast
- Inhomogeneous contrast
- Artifacts
Introduction to segmentation¶
Different types of segmentation¶
Basic segmentation: Applying a threshold to an image¶
Start out with a simple image of a cross with added noise
$$ I(x,y) = f(x,y) $$fig,ax = plt.subplots(1,2,figsize=(12,6))
nx = 5; ny = 5;
# Create the test image
xx, yy = np.meshgrid(np.arange(-nx, nx+1)/nx*2*np.pi, np.arange(-ny, ny+1)/ny*2*np.pi)
cross_im = 1.5*np.abs(np.cos(xx*yy))/(np.abs(xx*yy)+(3*np.pi/nx)) + np.random.uniform(-0.25, 0.25, size = xx.shape)
# Show it
im=ax[0].imshow(cross_im, cmap = 'hot'); ax[0].set_title("Image")
ax[1].hist(cross_im.ravel(),bins=10); ax[1].set_xlabel('Gray value'); ax[1].set_ylabel('Counts'); ax[1].set_title("Histogram");
Applying a threshold to an image¶
Applying the threshold is a deceptively simple operation
$$ I(x,y) = \begin{cases} 1, & f(x,y)\geq0.40 \\ 0, & f(x,y)<0.40 \end{cases}$$threshold = 0.4; thresh_img = cross_im > threshold
fig,ax = plt.subplots(1,2,figsize=(12,6))
ax[0].imshow(cross_im, cmap = 'hot', extent = [xx.min(), xx.max(), yy.min(), yy.max()]); ax[0].set_title("Image")
ax[0].plot(xx[np.where(thresh_img)]*0.9, yy[np.where(thresh_img)]*0.9,
'ks', markerfacecolor = 'green', alpha = 0.5,label = 'Threshold', markersize = 22); ax[0].legend(fontsize=12);
ax[1].hist(cross_im.ravel(),bins=10); ax[1].axvline(x=threshold,color='r',label='Threshold'); ax[1].legend(fontsize=12);
ax[1].set_xlabel('Gray value'); ax[1].set_ylabel('Counts'); ax[1].set_title("Histogram");
Noise and SNR¶
The noise in neutron imaging mainly originates from the amount of captured neutrons.
This noise is Poisson distributed and the signal to noise ratio is
$$SNR=\frac{E[x]}{s[x]}\sim\frac{N}{\sqrt{N}}=\sqrt{N}$$Problematic segmentation tasks¶

Woodland Encounter Bev Doolittle
Typical image features that makes life harder¶
In neutron imaging you see all these image phenomena.
Limited data problem¶
Different types of limited data:
- Few data points or limited amounts of images
- Unbalanced data
- Little or missing training data
Training data from NI is limited¶
- Long experiment times
- Few samples
- Some recycling from previous experiments is posible.
Augmentation to increase training data¶
Data augmentation is a method modify your exisiting data to obtain variations of it.
Augmentation will be used to increase the training data in the root segmenation example in the end of this lecture.
Simulation to increase training data¶
- Geometric models
- Template models
- Physical models
Both augmented and simulated data should be combined with real data.
Transfer learning¶
Transfer learning is a technique that uses a pre-trained network to
- Speed up training on your current data
- Support in cases of limited data
- Improve network performance
Unsupervised segmentation¶
Introducing clustering¶
test_pts = pd.DataFrame(make_blobs(n_samples=200, random_state=2018)[
0], columns=['x', 'y'])
plt.plot(test_pts.x, test_pts.y, 'r.');
k-means¶
The user only have to provide the number of classes the algorithm shall find.
Note The algorithm will find exactly the number you ask it to, it doesn't care if it makes sense!
Basic clustering example¶
N=3
fig, ax = plt.subplots(1,N,figsize=(18,4.5))
for i in range(N) :
km = KMeans(n_clusters=i+2, random_state=2018); n_grp = km.fit_predict(test_pts)
ax[i].scatter(test_pts.x, test_pts.y, c=n_grp)
ax[i].set_title('{0} groups'.format(i+2))
Add spatial information to k-means¶
orig = fits.getdata('../data/spots/mixture12_00001.fits')[::4,::4]
fig,ax = plt.subplots(1,6,figsize=(18,5)); x,y = np.meshgrid(np.linspace(0,1,orig.shape[0]),np.linspace(0,1,orig.shape[1]))
ax[0].imshow(orig, vmin=0, vmax=4000), ax[0].set_title('Original')
ax[1].imshow(x), ax[1].set_title('x-coordinates')
ax[2].imshow(y), ax[2].set_title('y-coordinates')
ax[3].imshow(flt.gaussian(orig, sigma=5)), ax[3].set_title('Weighted neighborhood')
ax[4].imshow(flt.sobel_h(orig),vmin=0, vmax=0.001),ax[4].set_title('Horizontal edges')
ax[5].imshow(flt.sobel_v(orig),vmin=0, vmax=0.001),ax[5].set_title('Vertical edges');
When can clustering be used on images?¶
- Single images
- Bimodal data
- Spectrum data
Clustering applied to wavelength resolved imaging¶
The data¶
tof = np.load('../data/tofdata.npy')
wtof = tof.mean(axis=2)
plt.imshow(wtof,cmap='gray');
plt.title('Average intensity all time bins');
Looking at the spectra¶
fig, ax= plt.subplots(1,2,figsize=(12,5))
ax[0].imshow(wtof,cmap='gray'); ax[0].set_title('Average intensity all time bins');
ax[0].plot(57,3,'ro'), ax[0].plot(15,30,'bo'), ax[0].plot(79,90,'go'); ax[0].plot(100,120,'co');
ax[1].plot(tof[30,15,:],'b', label='Sample'); ax[1].plot(tof[3,57,:],'r', label='Background'); ax[1].plot(tof[90,79,:],'g', label='Spacer'); ax[1].legend();ax[1].plot(tof[120,100,:],'c', label='Sample 2');
Reshaping¶
tofr=tof.reshape([tof.shape[0]*tof.shape[1],tof.shape[2]])
print("Input ToF dimensions",tof.shape)
print("Reshaped ToF data",tofr.shape)
Input ToF dimensions (128, 128, 661) Reshaped ToF data (16384, 661)
Setting up and running k-means¶
- We can clearly see that there is void on the sides of the specimens.
- There is also a separating band between the specimens.
- Finally we have to decide how many regions we want to find in the specimens. Let's start with two regions with different characteristics.
km = KMeans(n_clusters=4, random_state=2018) # Random state is an initialization parameter for the random number generator
c = km.fit_predict(tofr).reshape(tof.shape[:2]) # Label image
kc = km.cluster_centers_.transpose() # cluster centroid spectra
Results from the first try
fig,axes = plt.subplots(1,3,figsize=(18,5)); axes=axes.ravel()
axes[0].imshow(wtof,cmap='viridis'); axes[0].set_title('Average image')
p=axes[1].plot(kc); axes[1].set_title('Cluster centroid spectra'); axes[1].set_aspect(tof.shape[2], adjustable='box')
cmap=ps.buildCMap(p) # Create a color map with the same colors as the plot
im=axes[2].imshow(c,cmap=cmap); plt.colorbar(im);
axes[2].set_title('Cluster map');
plt.tight_layout()
We need more clusters¶
- Experiment data has variations on places we didn't expect k-means to detect as clusters.
- We need to increase the number of clusters!
Increasing the number of clusters¶
What happens when we increase the number of clusters to ten?
km = KMeans(n_clusters=10, random_state=2018)
c = km.fit_predict(tofr).reshape(tof.shape[:2]) # Label image
kc = km.cluster_centers_.transpose() # cluster centroid spectra
Results of k-means with ten clusters
fig,axes = plt.subplots(1,3,figsize=(18,5)); axes=axes.ravel()
axes[0].imshow(wtof,cmap='gray'); axes[0].set_title('Average image')
p=axes[1].plot(kc); axes[1].set_title('Cluster centroid spectra'); axes[1].set_aspect(tof.shape[2], adjustable='box')
cmap=ps.buildCMap(p) # Create a color map with the same colors as the plot
im=axes[2].imshow(c,cmap=cmap); plt.colorbar(im);
axes[2].set_title('Cluster map');
plt.tight_layout()
Interpreting the clusters¶
fig,axes = plt.subplots(1,1,figsize=(14,5));
plt.plot(kc); axes.set_title('Cluster centroid spectra');
axes.add_patch(Ellipse((0,0.62), width=30,height=0.55,fill=False,color='r')) #,axes.set_aspect(tof.shape[2], adjustable='box')
axes.add_patch(Ellipse((0,0.24), width=30,height=0.15,fill=False,color='cornflowerblue')),axes.set_aspect(tof.shape[2], adjustable='box');
Cleaning up the works space¶
del km, c, kc, tofr, tof
Supervised segmentation¶
- Training: Requires training data
- Verification: Requires verification data
- Inference: The images you want to segment
k nearest neighbors¶
Create example data for supervised segmentation¶
blob_data, blob_labels = make_blobs(n_samples=100, random_state=2018)
test_pts = pd.DataFrame(blob_data, columns=['x', 'y'])
test_pts['group_id'] = blob_labels
plt.scatter(test_pts.x, test_pts.y, c=test_pts.group_id, cmap='viridis');
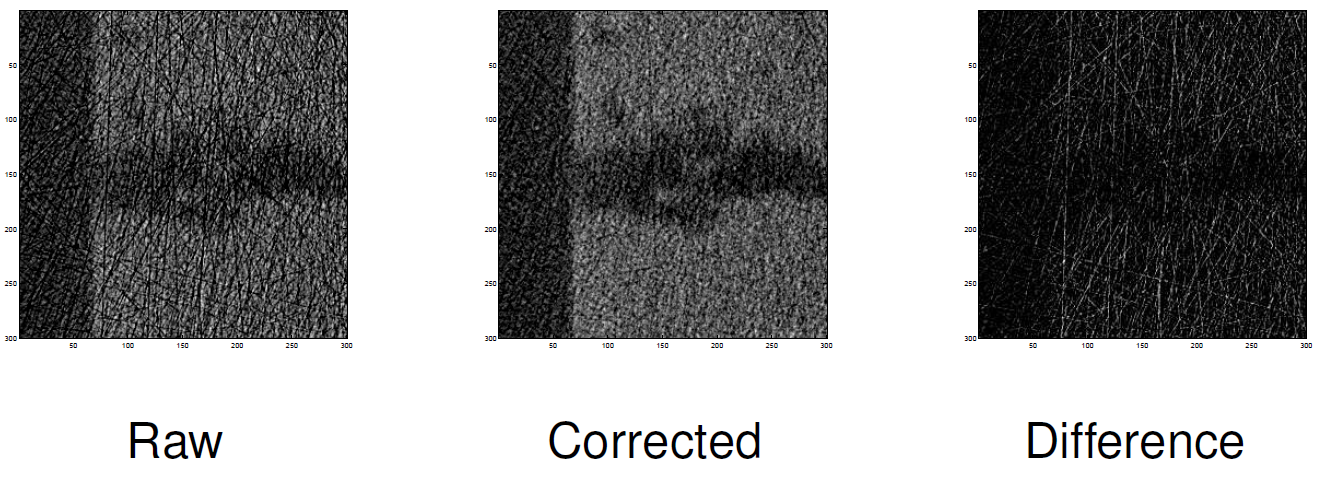
Detecting unwanted outliers in neutron images¶
orig= fits.getdata('../data/spots/mixture12_00001.fits')
annotated=io.imread('../data/spots/mixture12_00001.png'); mask=(annotated[:,:,1]==0)
r=600; c=600; w=256
ps.magnifyRegion(orig,[r,c,r+w,c+w],[15,7],vmin=400,vmax=4000,title='Neutron radiography')
Why are spots so relevant?¶
Marked-up spots¶
Baseline - Traditional spot cleaning algorithm¶
Parameters
- N Width of median filter.
- k Threshold level for outlier detection.
Bivariate histogram of the detection image¶
selem=np.ones([3,3])
forig=orig.astype('float32')
mimg = flt.median(forig,selem=selem)
d = np.abs(forig-mimg)
fig,ax=plt.subplots(1,2,figsize=(12,5))
h,x,y,u=ax[0].hist2d(forig.ravel(),d.ravel(), bins=100);
ax[0].set_xlabel('Input image - $f$'),ax[0].set_ylabel('$|f-med_{3x3}(f)|$'),ax[0].set_title('Bivariate histogram');
ax[1].imshow(np.log(h[:,::-1]+1).transpose(),vmin=0,vmax=10,extent=[x.min(),x.max(),y.min(),y.max()])
ax[1].set_xlabel('Input image - $f$'),ax[1].set_ylabel('$|f-med_{3x3}(f)|$'),ax[1].set_title('Log bivariate histogram');
The spot cleaning algorithm¶
def spotCleaner(img, threshold=0.95, selem=np.ones([3,3])) :
fimg=img.astype('float32')
mimg = flt.median(fimg,selem=selem)
timg = threshold < np.abs(fimg-mimg)
cleaned = mimg * timg + fimg * (1-timg)
return (cleaned,timg)
Testing the baseline algorithm for spot cleaning¶
baseclean,timg = spotCleaner(orig,threshold=1000)
ps.magnifyRegion(baseclean,[r,c,r+w,c+w],[12,3],vmin=400,vmax=4000,title='Cleaned image')
ps.magnifyRegion(timg,[r,c,r+w,c+w],[12,3],vmin=0,vmax=1,title='Detection image')
k nearest neighbors to detect spots¶
Prepare data¶
Training data
trainorig = forig[:,:1000].ravel()
traind = d[:,:1000].ravel()
trainmask = mask[:,:1000].ravel()
train_pts = pd.DataFrame({'orig': trainorig, 'd': traind, 'mask':trainmask})
Test data
testorig = forig[:,1000:].ravel()
testd = d[:,1000:].ravel()
testmask = mask[:,1000:].ravel()
test_pts = pd.DataFrame({'orig': testorig, 'd': testd, 'mask':testmask})
Train the model¶
k_class = KNeighborsClassifier(1)
k_class.fit(train_pts[['orig', 'd']], train_pts['mask'])
KNeighborsClassifier(n_neighbors=1)
Inspect decision space
xx, yy = np.meshgrid(np.linspace(test_pts.orig.min(), test_pts.orig.max(), 100),
np.linspace(test_pts.d.min(), test_pts.d.max(), 100),indexing='ij');
grid_pts = pd.DataFrame(dict(x=xx.ravel(), y=yy.ravel()))
grid_pts['predicted_id'] = k_class.predict(grid_pts[['x', 'y']])
plt.scatter(grid_pts.x, grid_pts.y, c=grid_pts.predicted_id, cmap='gray'); plt.title('Testing Points'); plt.axis('square');
Apply knn to unseen data¶
pred = k_class.predict(test_pts[['orig', 'd']])
pimg = pred.reshape(d[:,1000:].shape)
fig,ax = plt.subplots(1,3,figsize=(15,6))
ax[0].imshow(forig[:,1000:],vmin=0,vmax=4000), ax[0].set_title('Original image')
ax[1].imshow(pimg), ax[1].set_title('Predicted spot')
ax[2].imshow(mask[:,1000:]),ax[2].set_title('Annotated spots');
Performance check¶
ps.showHitMap(mask[:,1000:],timg[:,1000:])
plt.savefig('spotbaseline.png',dpi=300)
ps.showHitMap(mask[:,1000:], pimg)
plt.savefig('spotknn.png',dpi=300)
Some remarks about k-nn¶
- It takes quite some time to process
- You need to prepare training data
- Annotation takes time...
- Here we used the segmentation on the same type of image
- We should normalize the data
- This was a raw projection, what happens if we use a flat field corrected image?
- Finds more spots than baseline
- Data is very unbalanced, try a selection of non-spot data for training.
- Is it faster?
- Is there a drop segmentation performance?
Note There are other spot detection methods that perform better than the baseline.
Clean up¶
del k_class
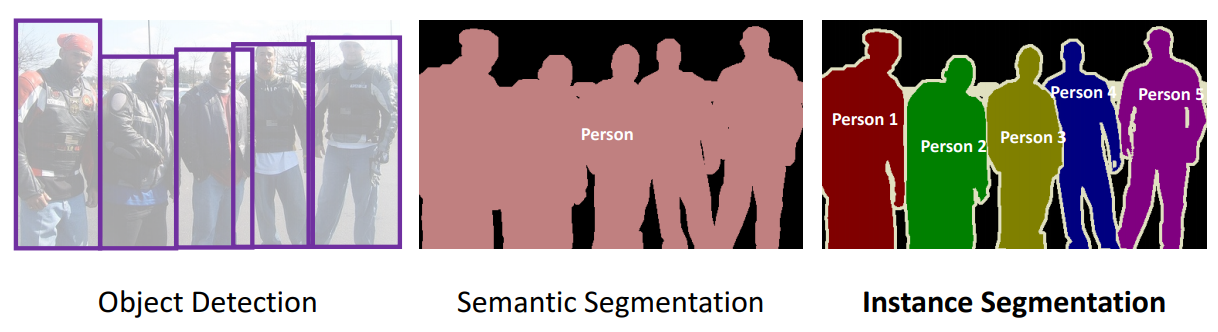
Convolutional neural networks for segmentation¶
The difference between classification and segmentation I¶
| Classification | Segmentation |
|---|---|
| pixels to classes | pixels to pixels |
 |
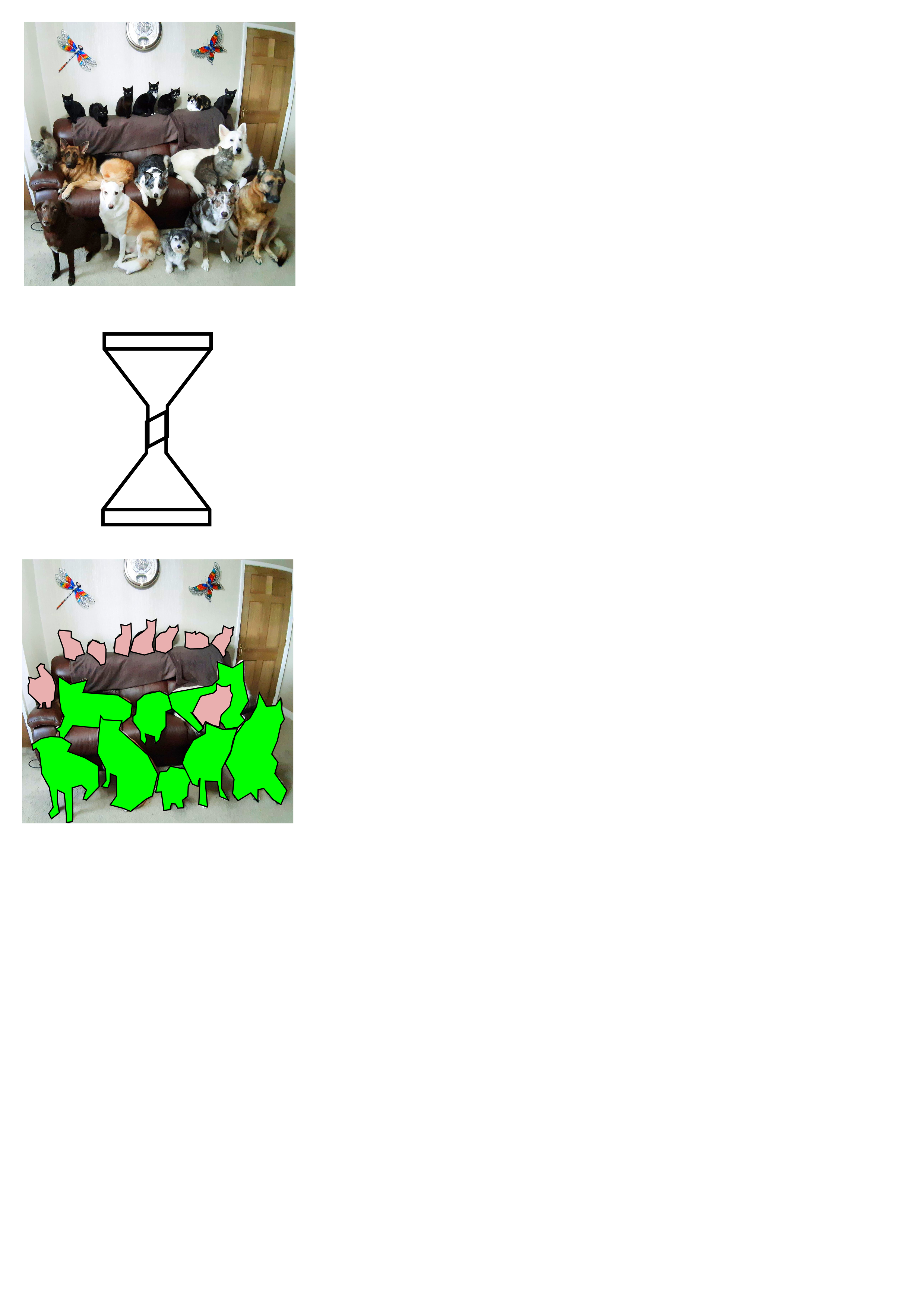 |
Different segmentation networks¶
Segmentation is mostly based on variations of the U-Net architechture
- AlexNET
- SegNET
- SegCaps
| Scales in traditional image processing | U-Net architecture |
|---|---|
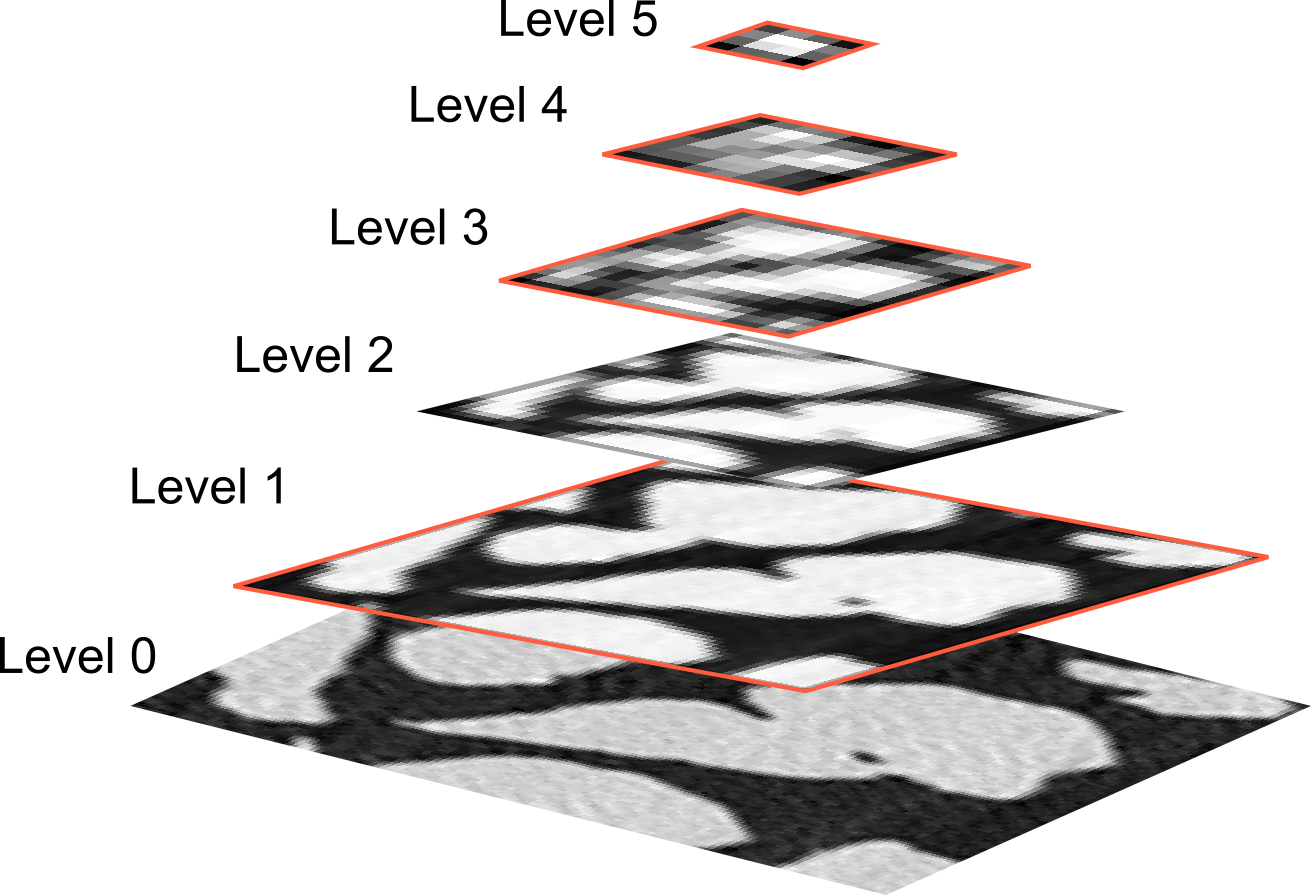 |
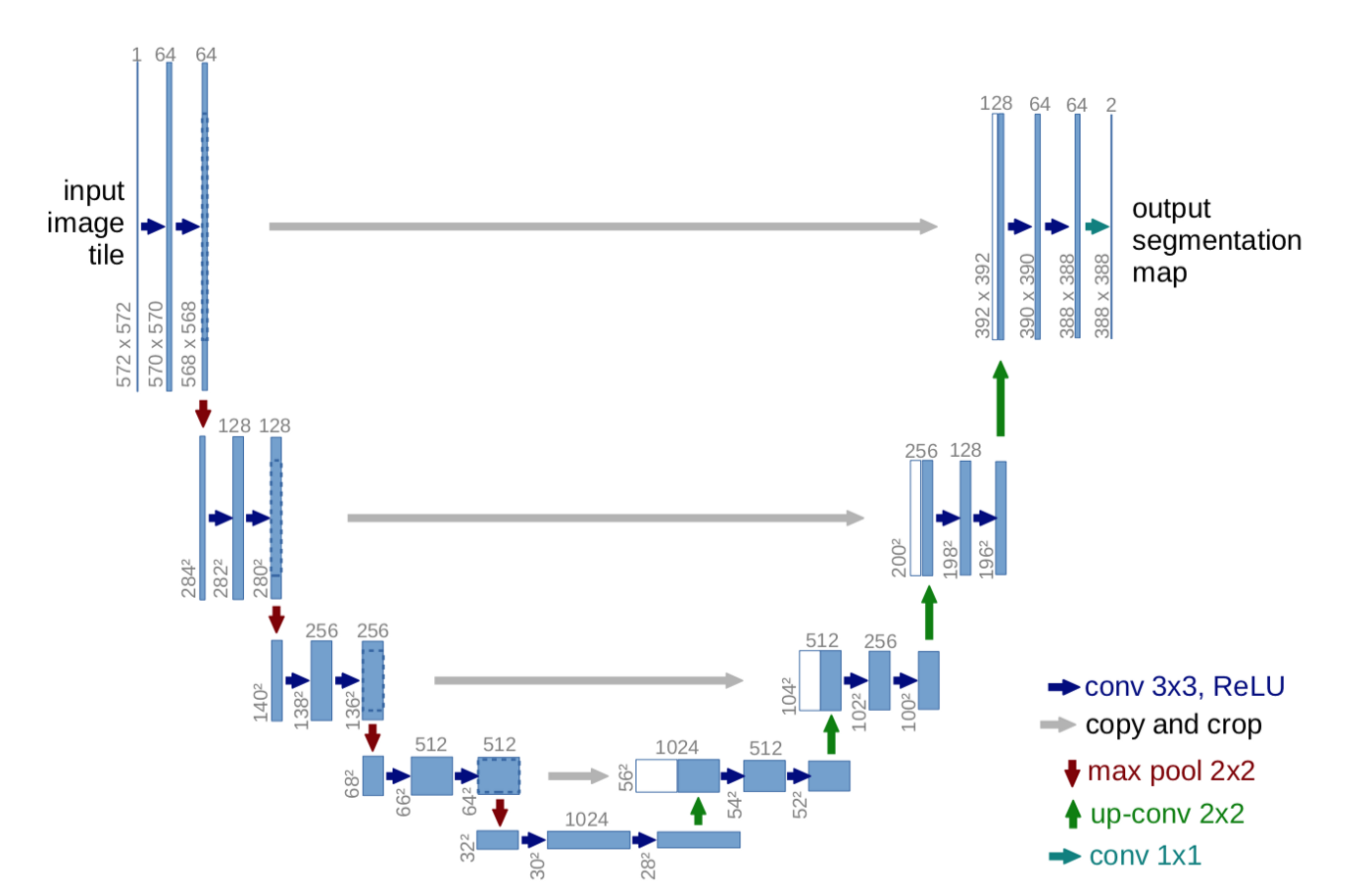 |
Training data¶
We have two choices:
- Use real data
- requires time consuming markup to provide training data
- corresponds to real life images
- Synthesize data
- flexible and provides both 'dirty' data and ground truth.
- model may not behave as real data
Preparing real data¶
We will use the spotty image as training data for this example
There is only one image!
fig,ax=plt.subplots(1,2,figsize=(12,5))
ax[0].imshow(forig,vmin=0,vmax=4000,cmap='gray'); ax[0].set_title('Original');
ax[1].imshow(mask,cmap='gray'); ax[1].set_title('Mask');
Prepare training, validation, and test data¶
Any analysis system must be verified to be demonstrate its performance and to further optimize it.
For this we need to split our data into three categories:
- Training data
- Test data
- Validation data
wpos = [1100,600]; ww = 512
train_img, valid_img, forigc = forig[128:256, 500:1300], forig[500:1000, 300:1500], forig[wpos[0]:(wpos[0]+ww),wpos[1]:(wpos[1]+ww)]
train_mask, valid_mask, maskc = mask[128:256, 500:1300], mask[500:1000, 300:1500], mask[wpos[0]:(wpos[0]+ww),wpos[1]:(wpos[1]+ww)]
fig, ax = plt.subplots(1, 4, figsize=(15, 6), dpi=300); ax=ax.ravel()
ax[0].imshow(train_img, cmap='bone',vmin=0,vmax=4000);ax[0].set_title('Train Image')
ax[1].imshow(train_mask, cmap='bone'); ax[1].set_title('Train Mask')
ax[2].imshow(valid_img, cmap='bone',vmin=0,vmax=4000); ax[2].set_title('Validation Image')
ax[3].imshow(valid_mask, cmap='bone');ax[3].set_title('Validation Mask');
| Training | Validation | Test |
|---|---|---|
| 70% | 15% | 15% |
Build a CNN for spot detection and cleaning¶
We need:
- Data
- Gray level image - our radiograph.
- Annotated image where the spots are marked.
- A U-net model
- Keras comes to our help
Build a U-Net model¶
def buildSpotUNet( base_depth = 48) :
in_img = Input((None, None, 1), name='Image_Input')
lay_1 = Conv2D(base_depth, kernel_size=(3, 3), padding='same',activation='relu')(in_img)
lay_2 = Conv2D(base_depth, kernel_size=(3, 3), padding='same',activation='relu')(lay_1)
lay_3 = MaxPooling2D(pool_size=(2, 2))(lay_2)
lay_4 = Conv2D(base_depth*2, kernel_size=(3, 3), padding='same',activation='relu')(lay_3)
lay_5 = Conv2D(base_depth*2, kernel_size=(3, 3), padding='same',activation='relu')(lay_4)
lay_6 = MaxPooling2D(pool_size=(2, 2))(lay_5)
lay_7 = Conv2D(base_depth*4, kernel_size=(3, 3), padding='same',activation='relu')(lay_6)
lay_8 = Conv2D(base_depth*4, kernel_size=(3, 3), padding='same',activation='relu')(lay_7)
lay_9 = UpSampling2D((2, 2))(lay_8)
lay_10 = concatenate([lay_5, lay_9])
lay_11 = Conv2D(base_depth*2, kernel_size=(3, 3), padding='same',activation='relu')(lay_10)
lay_12 = Conv2D(base_depth*2, kernel_size=(3, 3), padding='same',activation='relu')(lay_11)
lay_13 = UpSampling2D((2, 2))(lay_12)
lay_14 = concatenate([lay_2, lay_13])
lay_15 = Conv2D(base_depth, kernel_size=(3, 3), padding='same',activation='relu')(lay_14)
lay_16 = Conv2D(base_depth, kernel_size=(3, 3), padding='same',activation='relu')(lay_15)
lay_17 = Conv2D(1, kernel_size=(1, 1), padding='same',
activation='relu')(lay_16)
t_unet = Model(inputs=[in_img], outputs=[lay_17], name='SpotUNET')
return t_unet
Model summary
t_unet = buildSpotUNet(base_depth=24)
t_unet.summary()
WARNING:tensorflow:From /home/travis/miniconda/envs/book/lib/python3.7/site-packages/tensorflow_core/python/ops/resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.
Instructions for updating:
If using Keras pass *_constraint arguments to layers.
WARNING:tensorflow:From /home/travis/miniconda/envs/book/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:4070: The name tf.nn.max_pool is deprecated. Please use tf.nn.max_pool2d instead.
Model: "SpotUNET"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
Image_Input (InputLayer) (None, None, None, 1 0
__________________________________________________________________________________________________
conv2d_1 (Conv2D) (None, None, None, 2 240 Image_Input[0][0]
__________________________________________________________________________________________________
conv2d_2 (Conv2D) (None, None, None, 2 5208 conv2d_1[0][0]
__________________________________________________________________________________________________
max_pooling2d_1 (MaxPooling2D) (None, None, None, 2 0 conv2d_2[0][0]
__________________________________________________________________________________________________
conv2d_3 (Conv2D) (None, None, None, 4 10416 max_pooling2d_1[0][0]
__________________________________________________________________________________________________
conv2d_4 (Conv2D) (None, None, None, 4 20784 conv2d_3[0][0]
__________________________________________________________________________________________________
max_pooling2d_2 (MaxPooling2D) (None, None, None, 4 0 conv2d_4[0][0]
__________________________________________________________________________________________________
conv2d_5 (Conv2D) (None, None, None, 9 41568 max_pooling2d_2[0][0]
__________________________________________________________________________________________________
conv2d_6 (Conv2D) (None, None, None, 9 83040 conv2d_5[0][0]
__________________________________________________________________________________________________
up_sampling2d_1 (UpSampling2D) (None, None, None, 9 0 conv2d_6[0][0]
__________________________________________________________________________________________________
concatenate_1 (Concatenate) (None, None, None, 1 0 conv2d_4[0][0]
up_sampling2d_1[0][0]
__________________________________________________________________________________________________
conv2d_7 (Conv2D) (None, None, None, 4 62256 concatenate_1[0][0]
__________________________________________________________________________________________________
conv2d_8 (Conv2D) (None, None, None, 4 20784 conv2d_7[0][0]
__________________________________________________________________________________________________
up_sampling2d_2 (UpSampling2D) (None, None, None, 4 0 conv2d_8[0][0]
__________________________________________________________________________________________________
concatenate_2 (Concatenate) (None, None, None, 7 0 conv2d_2[0][0]
up_sampling2d_2[0][0]
__________________________________________________________________________________________________
conv2d_9 (Conv2D) (None, None, None, 2 15576 concatenate_2[0][0]
__________________________________________________________________________________________________
conv2d_10 (Conv2D) (None, None, None, 2 5208 conv2d_9[0][0]
__________________________________________________________________________________________________
conv2d_11 (Conv2D) (None, None, None, 1 25 conv2d_10[0][0]
==================================================================================================
Total params: 265,105
Trainable params: 265,105
Non-trainable params: 0
__________________________________________________________________________________________________
Functions to prepare data for training¶
def prep_img(x, n=1):
return (prep_mask(x, n=n)-train_img.mean())/train_img.std()
def prep_mask(x, n=1):
return np.stack([np.expand_dims(x, -1)]*n, 0)
Test the untrained model¶
- We can make predictions with an untrained model (default parameters)
- but we clearly do not expect them to be very good
unet_pred = t_unet.predict(prep_img(forigc))[0, :, :, 0]
WARNING:tensorflow:From /home/travis/miniconda/envs/book/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:422: The name tf.global_variables is deprecated. Please use tf.compat.v1.global_variables instead.
fig, m_axs = plt.subplots(2, 3, figsize=(15, 6), dpi=150)
for c_ax in m_axs.ravel():
c_ax.axis('off')
((ax1, _, ax2), (ax3, ax4, ax5)) = m_axs
ax1.imshow(train_img, cmap='bone',vmin=0,vmax=4000); ax1.set_title('Train Image')
ax2.imshow(train_mask, cmap='viridis'); ax2.set_title('Train Mask')
ax3.imshow(forigc, cmap='bone',vmin=0, vmax=4000); ax3.set_title('Test Image')
ax4.imshow(unet_pred, cmap='viridis', vmin=0, vmax=0.1); ax4.set_title('Predicted Segmentation')
ax5.imshow(maskc, cmap='viridis'); ax5.set_title('Ground Truth');
The untrained model doesn't perform very well. You clearly see that the image structures appear here. What is worth noting the spots already appear as amplified. This what we want to improve during the training.
Training conditions¶
- Loss function - Binary cross-correlation
- Optimizer - ADAM
- 20 Epochs (training iterations)
- Metrics
- True positives
- False positives
- True negatives
- False negatives
- Binary accuracy (percentage of pixels correct classified) $$BA=\frac{1}{N}\sum_i(f_i==g_i)$$
- Precision $$Precision=\frac{TP}{TP+FP}$$
- Recall $$Recall=\frac{TP}{TP+FN}$$
- Area under reciever operating characteristics (ROC) curve $$AUC=\int ROC$$
- Mean absolute error $$MAE=\frac{1}{N}\sum_i|f_i-g_i|$$
Compile the model¶
mlist = [
metrics.TruePositives(name='tp'), metrics.FalsePositives(name='fp'),
metrics.TrueNegatives(name='tn'), metrics.FalseNegatives(name='fn'),
metrics.BinaryAccuracy(name='accuracy'), metrics.Precision(name='precision'),
metrics.Recall(name='recall'), metrics.AUC(name='auc'),
metrics.MeanAbsoluteError(name='mae')]
t_unet.compile(
loss=loss.BinaryCrossentropy(), # we use the binary cross-entropy to optimize
optimizer=opt.Adam(lr=1e-3), # we use ADAM to optimize
metrics=mlist # we keep track of the metrics in mlist
)
WARNING:tensorflow:From /home/travis/miniconda/envs/book/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:3172: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version. Instructions for updating: Use tf.where in 2.0, which has the same broadcast rule as np.where
A general note on the following demo¶
This is a very bad way to train a model;
- the optimizer can be tweaked, e.g. the learning rate can be changed,
- the training and validation data should not come from the same sample (and definitely not the same measurement).
- a single image does not provide a good base for a general spot detection algorithm.
The goal is to be aware of these techniques and have a feeling for how they can work for complex problems.
Training the spot detection model¶
Nsamples = 3
Nepochs = 20
loss_history = t_unet.fit(prep_img(train_img, n=Nsamples),
prep_mask(train_mask, n=Nsamples),
validation_data=(prep_img(valid_img),
prep_mask(valid_mask)),
epochs=Nepochs,
verbose = 2)
Train on 3 samples, validate on 1 samples Epoch 1/20 - 10s - loss: 0.0885 - tp: 0.0000e+00 - fp: 0.0000e+00 - tn: 304656.0000 - fn: 2544.0000 - accuracy: 0.9917 - precision: 0.0000e+00 - recall: 0.0000e+00 - auc: 0.4037 - mae: 0.0407 - val_loss: 0.1122 - val_tp: 10.0000 - val_fp: 5.0000 - val_tn: 593511.0000 - val_fn: 6474.0000 - val_accuracy: 0.9892 - val_precision: 0.6667 - val_recall: 0.0015 - val_auc: 0.5409 - val_mae: 0.0206 Epoch 2/20 - 7s - loss: 0.0929 - tp: 6.0000 - fp: 0.0000e+00 - tn: 304656.0000 - fn: 2538.0000 - accuracy: 0.9917 - precision: 1.0000 - recall: 0.0024 - auc: 0.5248 - mae: 0.0159 - val_loss: 0.1100 - val_tp: 23.0000 - val_fp: 13.0000 - val_tn: 593503.0000 - val_fn: 6461.0000 - val_accuracy: 0.9892 - val_precision: 0.6389 - val_recall: 0.0035 - val_auc: 0.5923 - val_mae: 0.0363 Epoch 3/20 - 7s - loss: 0.0951 - tp: 12.0000 - fp: 0.0000e+00 - tn: 304656.0000 - fn: 2532.0000 - accuracy: 0.9918 - precision: 1.0000 - recall: 0.0047 - auc: 0.5822 - mae: 0.0280 - val_loss: 0.1010 - val_tp: 16.0000 - val_fp: 8.0000 - val_tn: 593508.0000 - val_fn: 6468.0000 - val_accuracy: 0.9892 - val_precision: 0.6667 - val_recall: 0.0025 - val_auc: 0.6288 - val_mae: 0.0215 Epoch 4/20 - 7s - loss: 0.0853 - tp: 6.0000 - fp: 0.0000e+00 - tn: 304656.0000 - fn: 2538.0000 - accuracy: 0.9917 - precision: 1.0000 - recall: 0.0024 - auc: 0.6159 - mae: 0.0165 - val_loss: 0.1214 - val_tp: 10.0000 - val_fp: 2.0000 - val_tn: 593514.0000 - val_fn: 6474.0000 - val_accuracy: 0.9892 - val_precision: 0.8333 - val_recall: 0.0015 - val_auc: 0.6416 - val_mae: 0.0108 Epoch 5/20 - 7s - loss: 0.0965 - tp: 3.0000 - fp: 0.0000e+00 - tn: 304656.0000 - fn: 2541.0000 - accuracy: 0.9917 - precision: 1.0000 - recall: 0.0012 - auc: 0.6263 - mae: 0.0083 - val_loss: 0.0993 - val_tp: 18.0000 - val_fp: 7.0000 - val_tn: 593509.0000 - val_fn: 6466.0000 - val_accuracy: 0.9892 - val_precision: 0.7200 - val_recall: 0.0028 - val_auc: 0.7299 - val_mae: 0.0112 Epoch 6/20 - 7s - loss: 0.0812 - tp: 6.0000 - fp: 0.0000e+00 - tn: 304656.0000 - fn: 2538.0000 - accuracy: 0.9917 - precision: 1.0000 - recall: 0.0024 - auc: 0.7084 - mae: 0.0086 - val_loss: 0.0779 - val_tp: 76.0000 - val_fp: 36.0000 - val_tn: 593480.0000 - val_fn: 6408.0000 - val_accuracy: 0.9892 - val_precision: 0.6786 - val_recall: 0.0117 - val_auc: 0.7385 - val_mae: 0.0248 Epoch 7/20 - 7s - loss: 0.0759 - tp: 36.0000 - fp: 6.0000 - tn: 304650.0000 - fn: 2508.0000 - accuracy: 0.9918 - precision: 0.8571 - recall: 0.0142 - auc: 0.6784 - mae: 0.0191 - val_loss: 0.0646 - val_tp: 162.0000 - val_fp: 123.0000 - val_tn: 593393.0000 - val_fn: 6322.0000 - val_accuracy: 0.9892 - val_precision: 0.5684 - val_recall: 0.0250 - val_auc: 0.8015 - val_mae: 0.0364 Epoch 8/20 - 7s - loss: 0.0564 - tp: 93.0000 - fp: 48.0000 - tn: 304608.0000 - fn: 2451.0000 - accuracy: 0.9919 - precision: 0.6596 - recall: 0.0366 - auc: 0.7700 - mae: 0.0281 - val_loss: 0.0788 - val_tp: 218.0000 - val_fp: 219.0000 - val_tn: 593297.0000 - val_fn: 6266.0000 - val_accuracy: 0.9892 - val_precision: 0.4989 - val_recall: 0.0336 - val_auc: 0.8566 - val_mae: 0.0613 Epoch 9/20 - 7s - loss: 0.0781 - tp: 147.0000 - fp: 93.0000 - tn: 304563.0000 - fn: 2397.0000 - accuracy: 0.9919 - precision: 0.6125 - recall: 0.0578 - auc: 0.8611 - mae: 0.0644 - val_loss: 0.0633 - val_tp: 191.0000 - val_fp: 127.0000 - val_tn: 593389.0000 - val_fn: 6293.0000 - val_accuracy: 0.9893 - val_precision: 0.6006 - val_recall: 0.0295 - val_auc: 0.8870 - val_mae: 0.0446 Epoch 10/20 - 7s - loss: 0.0600 - tp: 123.0000 - fp: 54.0000 - tn: 304602.0000 - fn: 2421.0000 - accuracy: 0.9919 - precision: 0.6949 - recall: 0.0483 - auc: 0.8944 - mae: 0.0457 - val_loss: 0.0478 - val_tp: 148.0000 - val_fp: 67.0000 - val_tn: 593449.0000 - val_fn: 6336.0000 - val_accuracy: 0.9893 - val_precision: 0.6884 - val_recall: 0.0228 - val_auc: 0.8967 - val_mae: 0.0232 Epoch 11/20 - 7s - loss: 0.0393 - tp: 90.0000 - fp: 33.0000 - tn: 304623.0000 - fn: 2454.0000 - accuracy: 0.9919 - precision: 0.7317 - recall: 0.0354 - auc: 0.8938 - mae: 0.0188 - val_loss: 0.0533 - val_tp: 108.0000 - val_fp: 32.0000 - val_tn: 593484.0000 - val_fn: 6376.0000 - val_accuracy: 0.9893 - val_precision: 0.7714 - val_recall: 0.0167 - val_auc: 0.8892 - val_mae: 0.0137 Epoch 12/20 - 7s - loss: 0.0496 - tp: 66.0000 - fp: 18.0000 - tn: 304638.0000 - fn: 2478.0000 - accuracy: 0.9919 - precision: 0.7857 - recall: 0.0259 - auc: 0.8444 - mae: 0.0106 - val_loss: 0.0441 - val_tp: 127.0000 - val_fp: 42.0000 - val_tn: 593474.0000 - val_fn: 6357.0000 - val_accuracy: 0.9893 - val_precision: 0.7515 - val_recall: 0.0196 - val_auc: 0.9295 - val_mae: 0.0159 Epoch 13/20 - 7s - loss: 0.0358 - tp: 84.0000 - fp: 27.0000 - tn: 304629.0000 - fn: 2460.0000 - accuracy: 0.9919 - precision: 0.7568 - recall: 0.0330 - auc: 0.9254 - mae: 0.0133 - val_loss: 0.0468 - val_tp: 136.0000 - val_fp: 47.0000 - val_tn: 593469.0000 - val_fn: 6348.0000 - val_accuracy: 0.9893 - val_precision: 0.7432 - val_recall: 0.0210 - val_auc: 0.8983 - val_mae: 0.0237 Epoch 14/20 - 7s - loss: 0.0433 - tp: 90.0000 - fp: 27.0000 - tn: 304629.0000 - fn: 2454.0000 - accuracy: 0.9919 - precision: 0.7692 - recall: 0.0354 - auc: 0.8614 - mae: 0.0264 - val_loss: 0.0529 - val_tp: 145.0000 - val_fp: 54.0000 - val_tn: 593462.0000 - val_fn: 6339.0000 - val_accuracy: 0.9893 - val_precision: 0.7286 - val_recall: 0.0224 - val_auc: 0.8489 - val_mae: 0.0311 Epoch 15/20 - 7s - loss: 0.0543 - tp: 93.0000 - fp: 33.0000 - tn: 304623.0000 - fn: 2451.0000 - accuracy: 0.9919 - precision: 0.7381 - recall: 0.0366 - auc: 0.7918 - mae: 0.0382 - val_loss: 0.0532 - val_tp: 142.0000 - val_fp: 50.0000 - val_tn: 593466.0000 - val_fn: 6342.0000 - val_accuracy: 0.9893 - val_precision: 0.7396 - val_recall: 0.0219 - val_auc: 0.8291 - val_mae: 0.0302 Epoch 16/20 - 7s - loss: 0.0549 - tp: 93.0000 - fp: 33.0000 - tn: 304623.0000 - fn: 2451.0000 - accuracy: 0.9919 - precision: 0.7381 - recall: 0.0366 - auc: 0.7748 - mae: 0.0383 - val_loss: 0.0525 - val_tp: 129.0000 - val_fp: 41.0000 - val_tn: 593475.0000 - val_fn: 6355.0000 - val_accuracy: 0.9893 - val_precision: 0.7588 - val_recall: 0.0199 - val_auc: 0.8227 - val_mae: 0.0259 Epoch 17/20 - 7s - loss: 0.0509 - tp: 78.0000 - fp: 27.0000 - tn: 304629.0000 - fn: 2466.0000 - accuracy: 0.9919 - precision: 0.7429 - recall: 0.0307 - auc: 0.7772 - mae: 0.0324 - val_loss: 0.0515 - val_tp: 121.0000 - val_fp: 34.0000 - val_tn: 593482.0000 - val_fn: 6363.0000 - val_accuracy: 0.9893 - val_precision: 0.7806 - val_recall: 0.0187 - val_auc: 0.8337 - val_mae: 0.0205 Epoch 18/20 - 7s - loss: 0.0453 - tp: 75.0000 - fp: 21.0000 - tn: 304635.0000 - fn: 2469.0000 - accuracy: 0.9919 - precision: 0.7812 - recall: 0.0295 - auc: 0.8012 - mae: 0.0242 - val_loss: 0.0528 - val_tp: 112.0000 - val_fp: 28.0000 - val_tn: 593488.0000 - val_fn: 6372.0000 - val_accuracy: 0.9893 - val_precision: 0.8000 - val_recall: 0.0173 - val_auc: 0.8621 - val_mae: 0.0147 Epoch 19/20 - 7s - loss: 0.0414 - tp: 66.0000 - fp: 15.0000 - tn: 304641.0000 - fn: 2478.0000 - accuracy: 0.9919 - precision: 0.8148 - recall: 0.0259 - auc: 0.8439 - mae: 0.0150 - val_loss: 0.0555 - val_tp: 107.0000 - val_fp: 26.0000 - val_tn: 593490.0000 - val_fn: 6377.0000 - val_accuracy: 0.9893 - val_precision: 0.8045 - val_recall: 0.0165 - val_auc: 0.8866 - val_mae: 0.0114 Epoch 20/20 - 7s - loss: 0.0403 - tp: 66.0000 - fp: 15.0000 - tn: 304641.0000 - fn: 2478.0000 - accuracy: 0.9919 - precision: 0.8148 - recall: 0.0259 - auc: 0.8855 - mae: 0.0091 - val_loss: 0.0545 - val_tp: 118.0000 - val_fp: 27.0000 - val_tn: 593489.0000 - val_fn: 6366.0000 - val_accuracy: 0.9893 - val_precision: 0.8138 - val_recall: 0.0182 - val_auc: 0.8903 - val_mae: 0.0117
Training history plots¶
titleDict = {'tp': "True Positives",'fp': "False Positives",'tn': "True Negatives",'fn': "False Negatives", 'accuracy':"BinaryAccuracy",'precision': "Precision",'recall':"Recall",'auc': "Area under Curve", 'mae': "Mean absolute error"}
fig,ax = plt.subplots(2,5, figsize=(20,8), dpi=300)
ax =ax.ravel()
for idx,key in enumerate(titleDict.keys()):
ax[idx].plot(loss_history.epoch, loss_history.history[key], color='coral', label='Training')
ax[idx].plot(loss_history.epoch, loss_history.history['val_'+key], color='cornflowerblue', label='Validation')
ax[idx].set_title(titleDict[key]);
ax[9].axis('off');
axLine, axLabel = ax[0].get_legend_handles_labels() # Take the lables and plot line information from the first panel
lines =[]; labels = []; lines.extend(axLine); labels.extend(axLabel);fig.legend(lines, labels, bbox_to_anchor=(0.7, 0.3), loc='upper left');
Prediction on the training data¶
unet_train_pred = t_unet.predict(prep_img(train_img[:,wpos[1]:(wpos[1]+ww)]))[0, :, :, 0]
fig, m_axs = plt.subplots(1, 3, figsize=(18, 4), dpi=150); m_axs= m_axs.ravel();
for c_ax in m_axs: c_ax.axis('off')
m_axs[0].imshow(train_img[:,wpos[1]:(wpos[1]+ww)], cmap='bone', vmin=0, vmax=4000), m_axs[0].set_title('Train Image')
m_axs[1].imshow(unet_train_pred, cmap='viridis', vmin=0, vmax=0.2), m_axs[1].set_title('Predicted Training')
m_axs[2].imshow(train_mask[:,wpos[1]:(wpos[1]+ww)], cmap='viridis'), m_axs[2].set_title('Train Mask');
Prediction using unseen data¶
unet_pred = t_unet.predict(prep_img(forigc))[0, :, :, 0]
fig, m_axs = plt.subplots(1, 3, figsize=(18, 4), dpi=150); m_axs = m_axs.ravel() ;
for c_ax in m_axs: c_ax.axis('off')
m_axs[0].imshow(forigc, cmap='bone', vmin=0, vmax=4000); m_axs[0].set_title('Full Image')
f1=m_axs[1].imshow(unet_pred, cmap='viridis', vmin=0, vmax=0.1); m_axs[1].set_title('Predicted Segmentation'); fig.colorbar(f1,ax=m_axs[1]);
m_axs[2].imshow(maskc,cmap='viridis'); m_axs[2].set_title('Ground Truth');
Converting predictions to segments¶
fig, ax = plt.subplots(1,2, figsize=(12,4))
ax0=ax[0].imshow(unet_pred, vmin=0, vmax=0.1); ax[0].set_title('Predicted segmentation'); fig.colorbar(ax0,ax=ax[0])
ax[1].imshow(0.05<unet_pred), ax[1].set_title('Final segmenation');
Hit cases¶
gt = maskc
pr = 0.05<unet_pred
ps.showHitCases(gt,pr,cmap='gray')
Hit map¶
ps.showHitMap(gt,pr)
plt.savefig('spotunet.png')
Comparing the performance of the spot detection methods¶
Baseline

k-NN

U-Net

Concluding remarks about the spot detection¶
- Spot detection seems to be working well using the U-Net.
- A great amount of the spots are found.
- There are many false positive pixels - usually in the neighborhood of a spot.
- Some misclasifications are probably related to the annotation of the training image.
- Wide spot items may be related to the network depth.
- The demo sample is smooth, we didn't test the performance near edges
Improvements
- Increase the number of epochs in the training
- Increase the training data
- Add images with different SNR (real and simulated)
- Add images with different characteristics
Segmenting root networks in the rhizosphere¶
Background¶
- Soil and in particular the rhizosphere are of central interest for neutron imaging users.
- The experiments aim to follow the water distribution near the roots.
- The roots must be identified in 2D and 3D data
Today: much of this mark-up is done manually!

Acknowledgement: This work was done by Gian Guido Parenza as a master project.
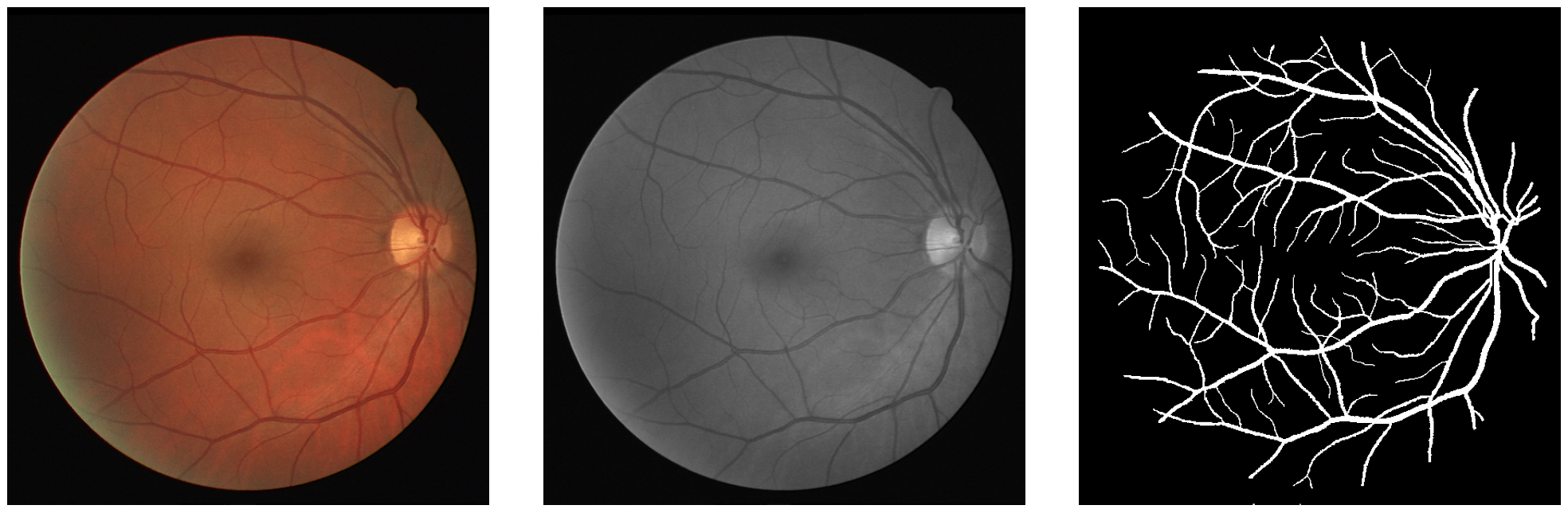
Available data¶
| Radiography | Tomography |
|---|---|
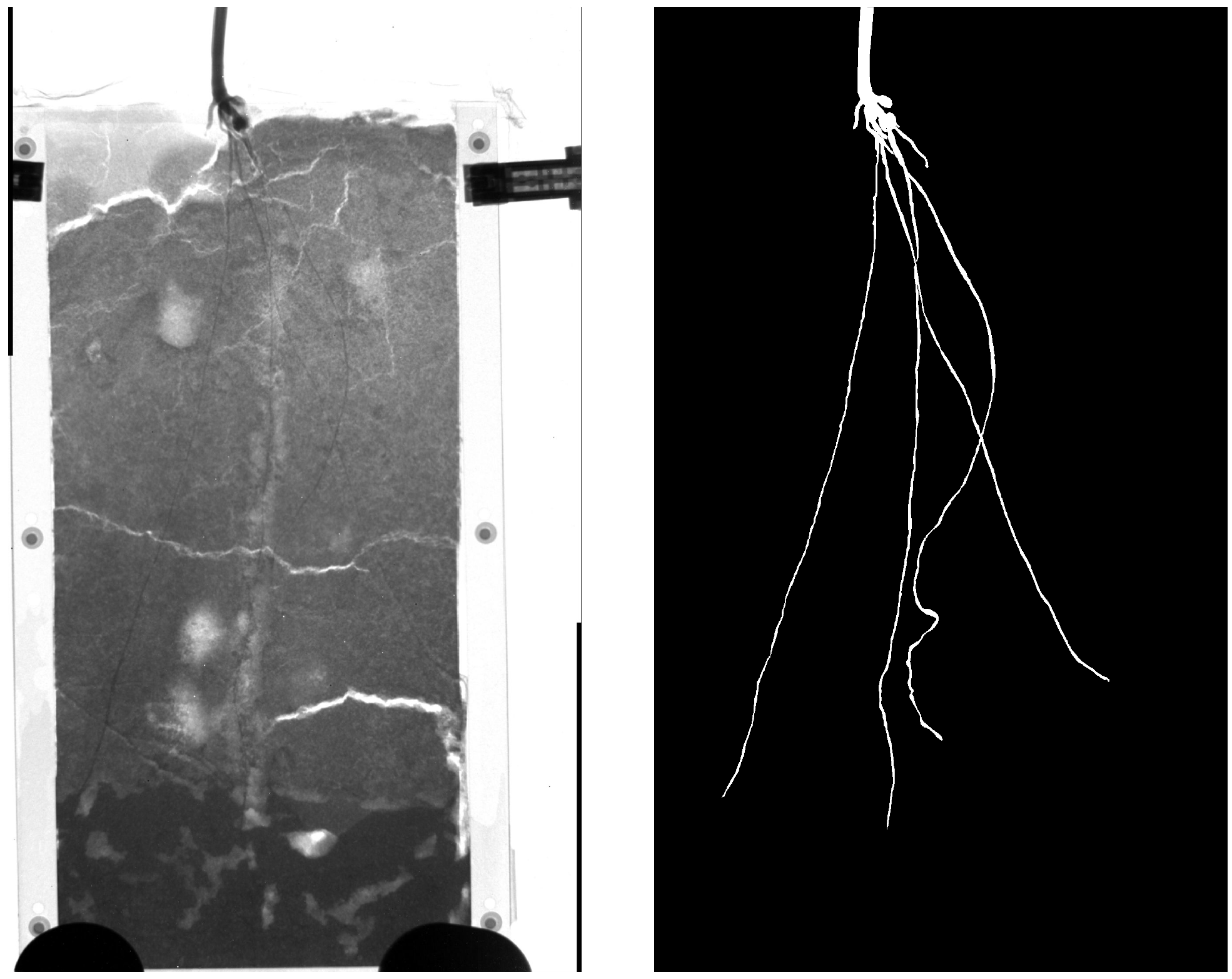 |
 |
| Provided by A. Carminati et al. | Provided by M. Menon et al. |
Results using current method¶
| Radiograph of a rhizobox | Current segmentation |
|---|---|
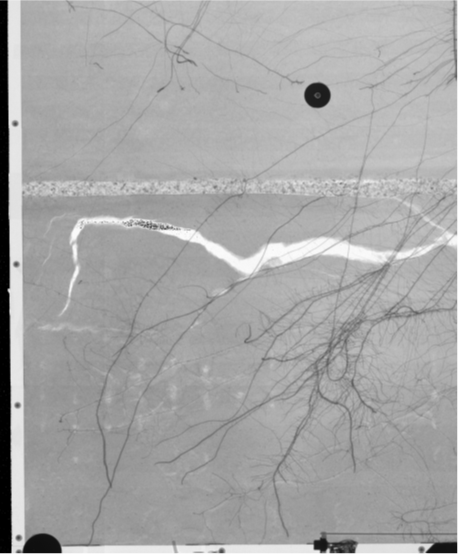 |
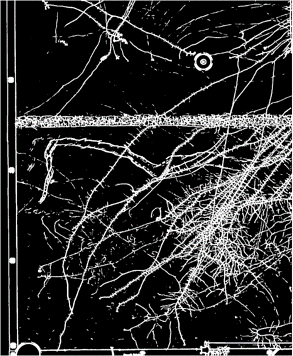 |
Problems: Unwanted elements are marked as roots
- Elements of the container
- Soil cracks
- The porous barrier
Workflow¶
Loss functions¶
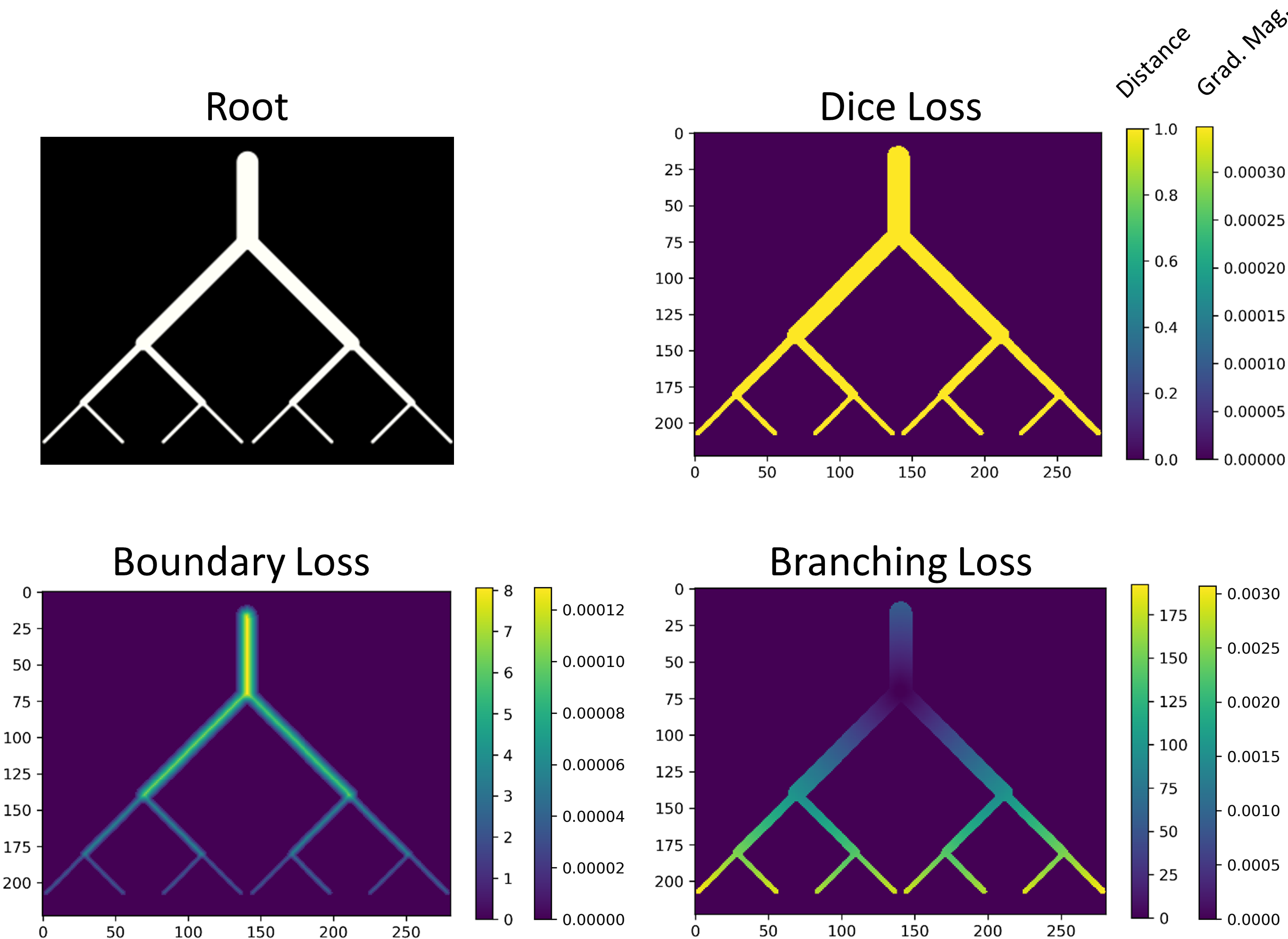
The impact of different loss functions¶
| Compare different loss functions | Details of branching loss |
|---|---|
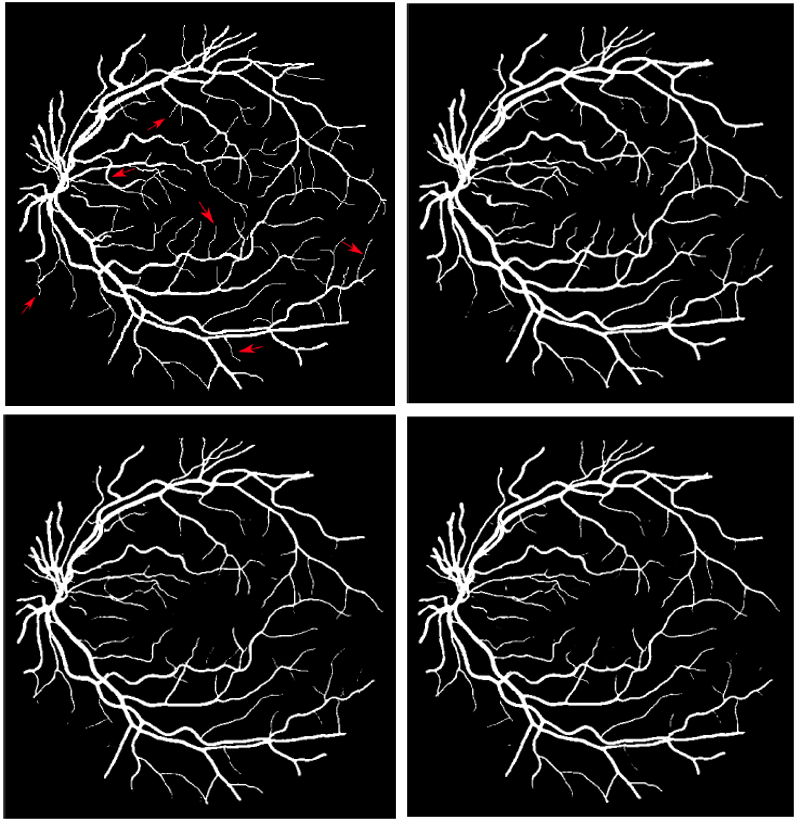 |
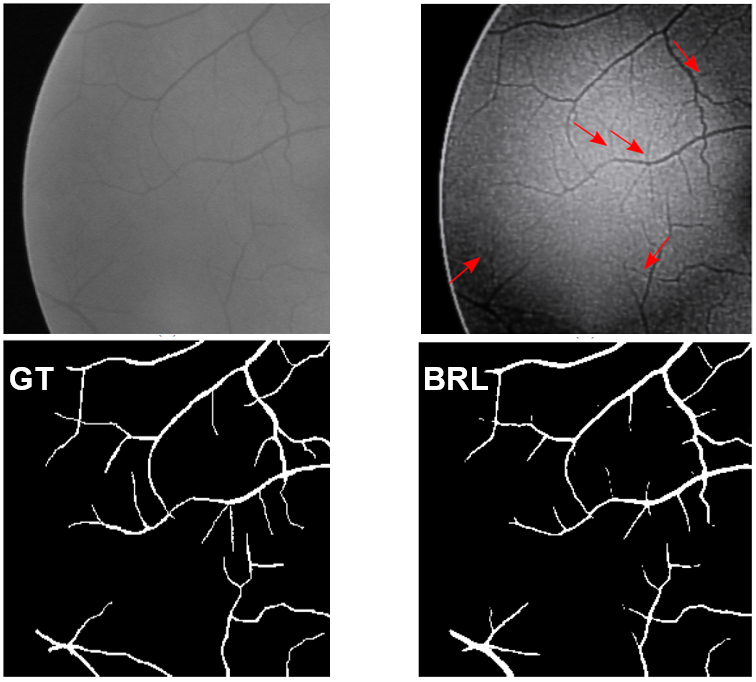 |
Transfer learning¶
- We have little available annotated neutron data
- There is more radiographs than tomograms
Our options
- Simulations using root network simulators and Monte Carlo neutron simulation
- Use data with similar features.
Medical image processing is the saviour!
Trying transfer learning on the roots¶
| U-Net trained with roots only | U-Net trained with transfer learning |
|---|---|
 |
 |
Transfer learning
- Speeds up the training with root data
- Improves the segmented results
- Can even be used for 3D data
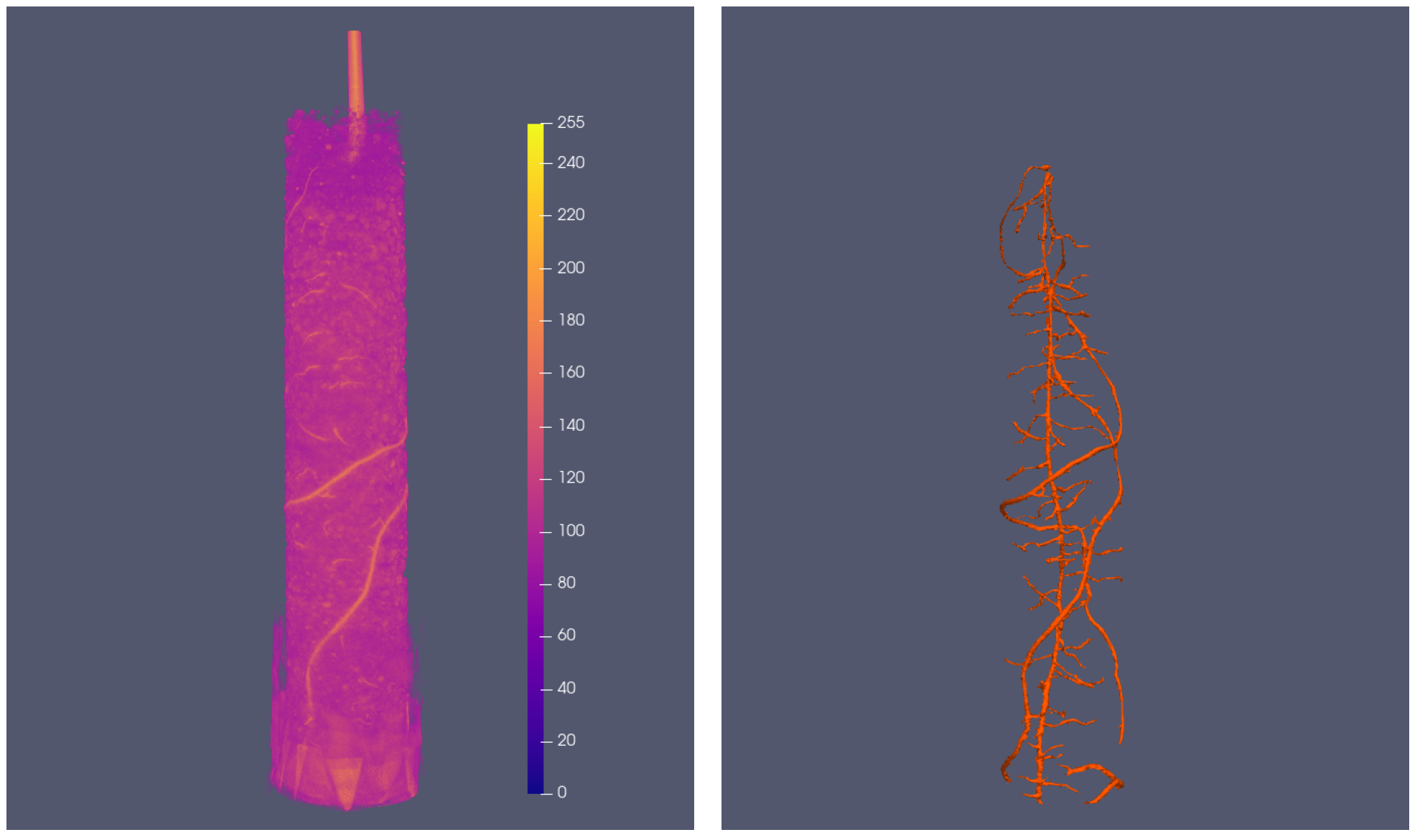
The model can also be used for volume data¶
after some modification
| Original tomography data and ground truth | Segmentations |
|---|---|
 |
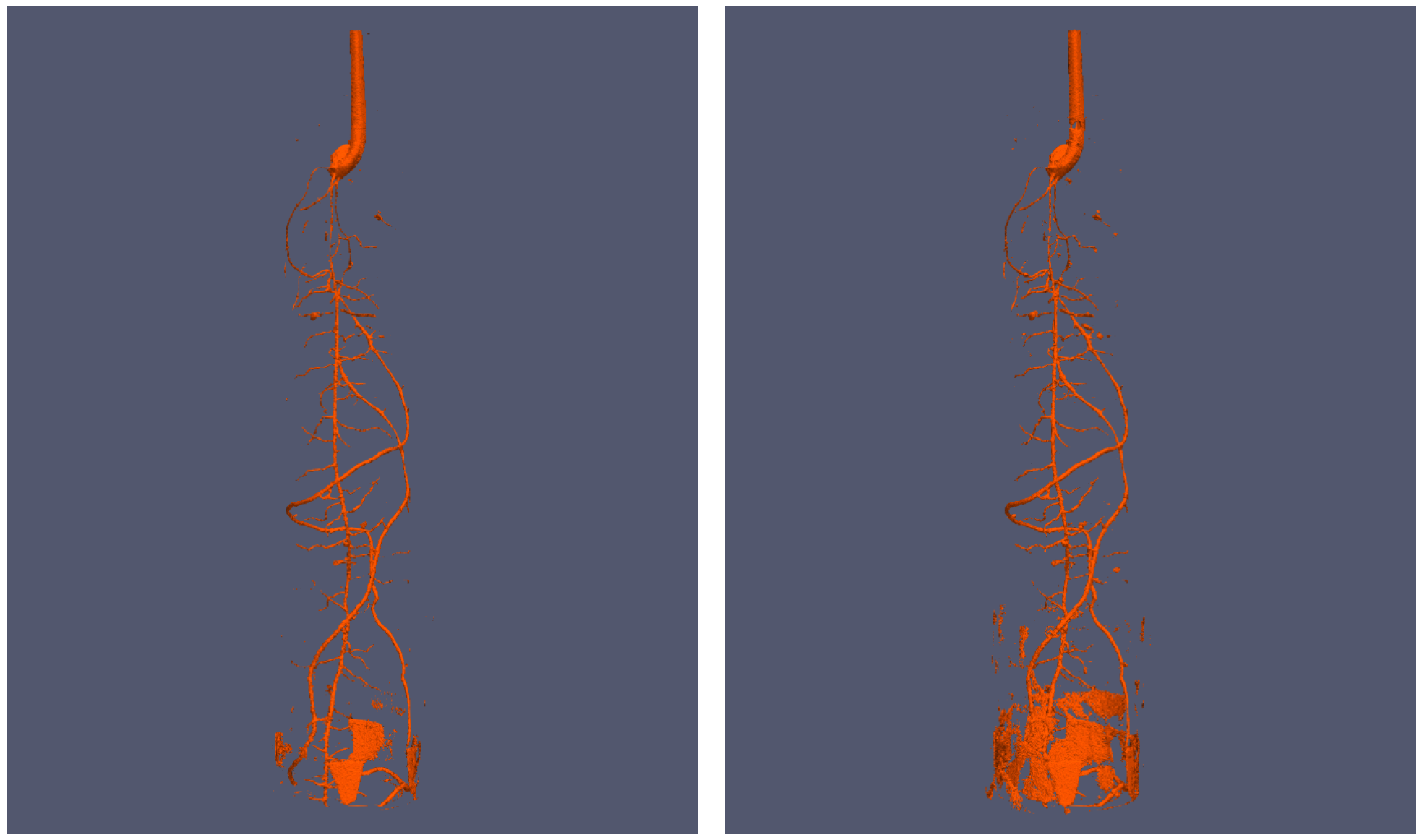 |
Summary¶
This project has shown that
- Convolutional NNs can segment roots in soil
- Will save a lot of work in future rhizospere experiment
The models still need more training to cover wider variations in the data.
Concluding remarks¶
We have demonstrated how some machine learning techniques can be used on neutron images:
- Some background to the segmentation problem and neutron imaging
- k-means - to segment ToF spectra
- Spot detection
- Baseline algorithm
- k-Nearest neighbors
- U-Net
- Root segmentation
- U-Net
- Different loss metrics
- Training performance